Nguyen, H. L. & Marohn, J. A.“Reverse Monte Carlo Reconstruction of Electron Spin-label Coordinates from Scanned-probe Magnetic Resonance Microscope Signals”, submitted 20 February 2018 [arXiv:1802.07247]. The abstract of the paper reads
Individual electron spins have been observed using magnetic resonance in combination with a number of distinct detection approaches. The coordinates of an individual electron spin can then in principle be determined by introducing a 10 to 100 nm diameter magnetic needle, scanning the needle, and collecting signal as a function of the needle’s position. Although individual electrons have recently been localized with nanometer precision in this way using a nitrogen-vacancy center in diamond as the spin detector, the experiment’s low signal-to-noise ratio limited acquisition to two-dimensional scanning of just a few dozen data points and was incompatible with nitroxide spin labels widely used to label proteins and nucleic acids.
We introduce and numerically simulate a protocol for detecting and imaging individual nitroxide electron spins mechanically with high spatial resolution. In our protocol a scanned magnet-tipped cantilever is brought near the sample, modulated microwaves are applied to resonantly excite electron spins, and changes in spin magnetization are detected as a shift in the mechanical frequency of the cantilever. By carefully applying resonant microwaves in short bursts in synchrony with the cantilever’s oscillation, we propose to retain high spatial resolution even at large cantilever amplitude where sensitivity is highest. Numerical simulations reveal nanometer-diameter rings of frequency-shift signal as the tip is scanned. Our primary finding is that it is possible — using a Bayesian, reverse Monte Carlo algorithm introduced here — to obtain the full three-dimensional distribution of electron coordinates from the signal rings revealed in a two-dimensional frequency-shift map. This reduction in dimensionality brings within reach, on a practical timescale, the angstrom-resolution three-dimensional imaging of spin-labeled macromolecules.
Our main findings are summarized pretty well in Figure 5 of the paper, reproduced below.
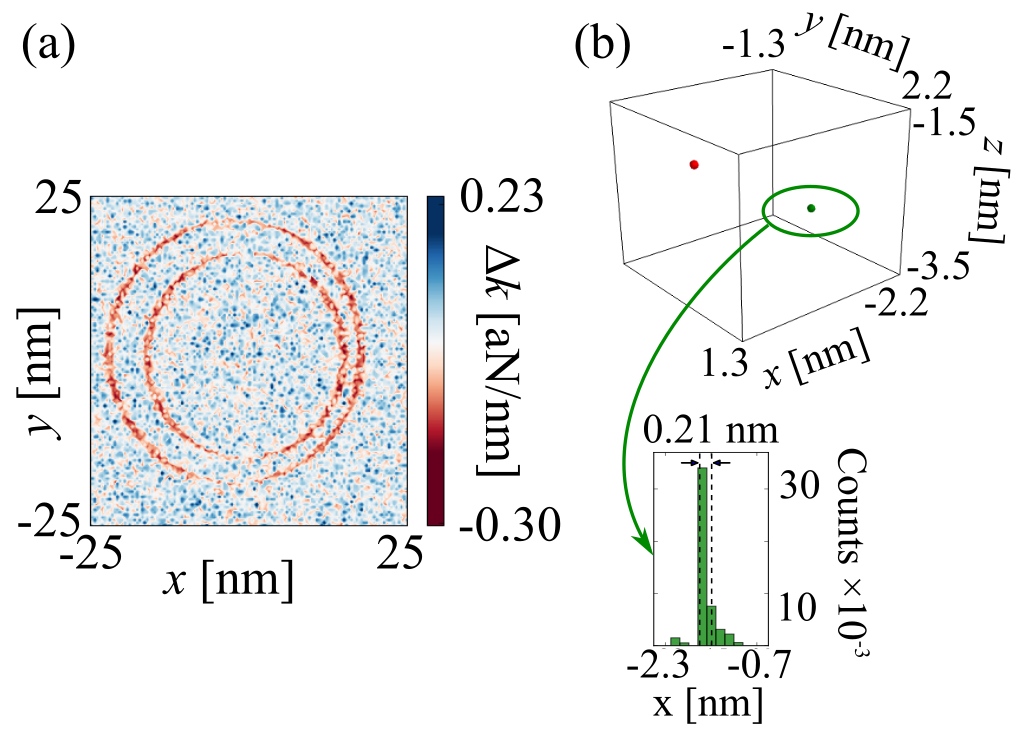
(a) Simulated 2D scanned force-gradient signal for the doubly spin-labeled T4 lysozyme mutant pdb 3K2R. (b) Image reconstructed from the simulated signal using the Bayesian Markov chain Monte-Carlo approach discussed in the text. (Upper) Reconstructed three-dimensional spin density showing the location of two individual electron spins separated by 21 Å. (Lower) The posterior distribution of the x-position of one of the two electron spins, showing a resolution of 2.1 Å.
This work was funded by the U.S. Army Research Office.